400 Dowman Dr.
Math and Science Center
Room: N246
Phone: 404-727-4930
Email: lfinzi@emory.edu
Tethered Particle Motion technique, or Tethered Particle Microscopy, (TPM)
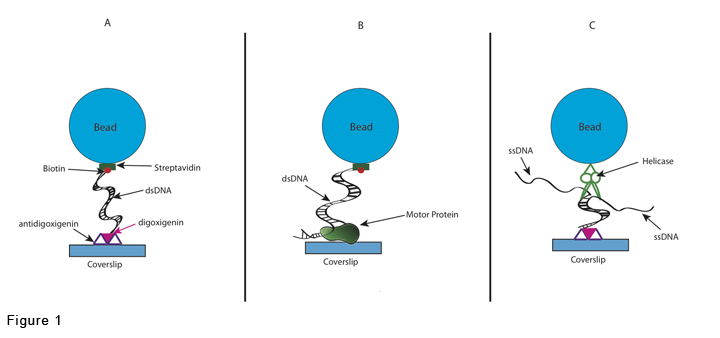
There are three main TPM experimental setups which are described in Figure 1.
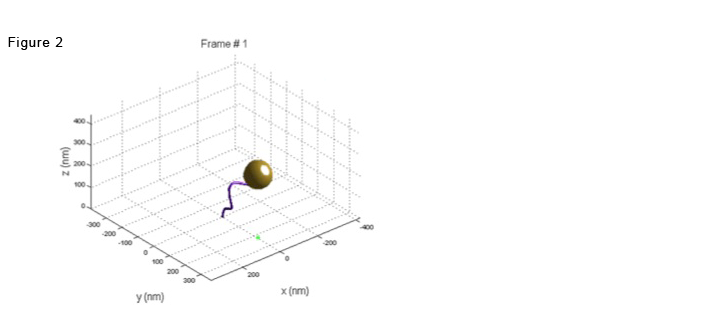
In the most common of them, a submicron diameter bead is tethered by a single DNA molecule to a glass surface. The bead exhibits a Brownian motion restricted by the DNA tether. See figure 2:
and movie 1:
Black crosses on lower bead are tracking results
If the DNA tether undergoes topological changes that alter its effective length, the Brownian excursions of the bead, measured as the projected displacement vector, will change correspondingly.
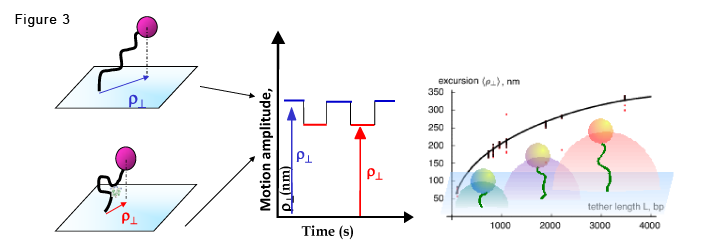
In the figure, the DNA is represented by a black curved line. If a protein (green complex) induces looping, the bead's tether shortens and reduces the bead's amplitude of motion (excursion).When the DNA molecule changes conformation, due to protein association and dissociation (left), the amplitude of the Brownian motion, rho, will fluctuate, in time between two levels, resembling a telegraphic signal (right).
Our group has also used TPM to study the diffusion of membrane proteins (pub. 6 in Methods).
Follow the link to Research/Methods/TPM to see how we improved this technique.